Scientists have used computer modelling to identify potential ‘vulnerable sites’ on a key protein found in coronavirus – paving the way for possible new drug treatments in the future.
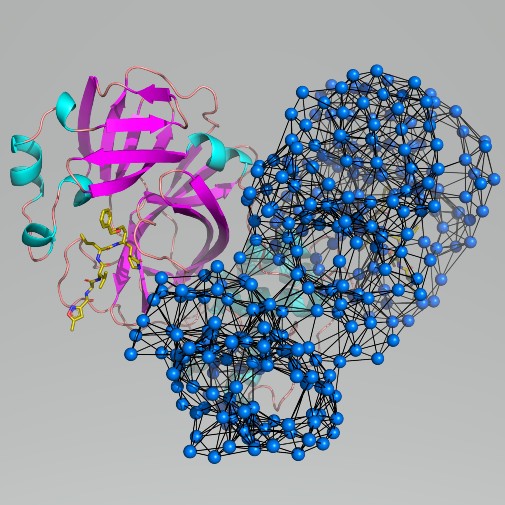
The coronavirus responsible for the Covid-19 epidemic deploys dozens of viral biomolecules when it invades host cells with the disease. One of these is a compact protein, the ’main protease’, whose function is critical to the virus.
Scientists at the University of York analysed the structure of the protease by computational modelling – simulating the protein’s motions.
The model is very efficient because it works by developing a bird’s eye view of the protein structure. It suggests sites on the protein that may be accessible to new drugs.
The technique could be used for spotting ‘vulnerable’ sites for inhibition in other proteins, including other SARS-CoV-2 proteins.
Prof McLeish said the research was not related to the current vaccines, which are based on the ’Spike’ protein, but is a study of another key protein in the Covid process.
He added: “It is more relevant to potential future drugs than to future vaccines, as the motions of the protein that it uncovers point to new ’sites’ on the protein where binding small molecules might disrupt the protein function.
“The advantage of these sites, and our method in general, is that they are not the ‘obvious’ ones that compete with the normal binding of the protein, but other sites that can be accessed even when the usual binding sites are occupied.”
Igors Dubanevics added: “We have identified promising druggable sites in the main protease via computer simulations and some of them have been supported by the newest studies by other groups.
“The next logical step would be to investigate the identified sites by conducting biological experiments in a lab.